 |
 |
AbstractPurposeThe probability of recurrence of cancer after adjuvant or definitive radiotherapy in patients with human papillomavirus-negative (HPV(–)) head and neck squamous cell carcinoma (HNSCC) varies for each patient. This study aimed to identify and validate radiation sensitivity signature (RSS) of patients with HPV(–) HNSCC to predict the recurrence of cancer after radiotherapy.
Materials and MethodsClonogenic survival assays were performed to assess radiosensitivity in 14 HNSCC cell lines. We identified genes closely correlated with radiosensitivity and validated them in The Cancer Genome Atlas (TCGA) cohort. The validated RSS were analyzed by ingenuity pathway analysis (IPA) to identify canonical pathways, upstream regulators, diseases and functions, and gene networks related to radiosensitive genes in HPV(–) HNSCC.
ResultsThe survival fraction of 14 HNSCC cell lines after exposure to 2 Gy of radiation ranged from 48% to 72%. Six genes were positively correlated and 35 genes were negatively correlated with radioresistance, respectively. RSS was validated in the HPV(–) TCGA HNSCC cohort (n = 203), and recurrence-free survival (RFS) rate was found to be significantly lower in the radioresistant group than in the radiosensitive group (p = 0.035). Cell death and survival, cell-to-cell signaling, and cellular movement were significantly enriched in RSS, and RSSs were highly correlated with each other.
IntroductionHead and neck squamous cell carcinoma (HNSCC) arises in the mucosal lining of oral cavity, oropharynx, larynx, or hypopharynx, and is the sixth most common cancer worldwide [1]. HNSCC represents about 6% of all cancer cases and accounts for an estimated 650,000 new cases and 350,000 deaths worldwide every year [2]. About one-third of patients are diagnosed with early-stage of HNSCC, whereas a typical patient is diagnosed with advanced stage of HNSCC with lymph node metastases [3].
Early-stage HNSCC is treated by surgery or radiation, and advanced-stage HNSCC commonly requires combined therapy such as surgery, radiotherapy, and chemotherapy. Despite of the improved quality of life in patients with HNSCC after advancement in treatment modality such as advanced surgical technique [4] and radiotherapy [5], survival rates have not markedly improved in recent decades [6]. In particular, the intensity-modulated radiotherapy (IMRT; a three-dimensional modern radiotherapy technique) allowed sculpting of the radiation dose to the target volume and resulted in significantly reduced late toxicities compared to that in older radiation techniques, but showed no difference in locoregional recurrence and disease-free and overall survival in most cases when compared to those in the older radiation techniques [7].
Recently, besides alcohol consumption and tobacco history, which are well-known risk factors for the development of HNSCC, infection with the human papillomavirus (HPV) has been identified as an independent parameter in the development of HNSCC [8]. Patients with HPV-positive (HPV(+)) HNSCC might have a different etiology and favorable prognosis compared to patients with HPV-negative (HPV(–)) HNSCC [9]. In a multicenter study, patients with HPV(+) HNSCC showed better prognosis in locoregional recurrence and overall survival than patients with HPV(–) HNSCC [10]. In addition, patients with HPV(+) HNSCC were more sensitive to radiation than HPV(–) HNSCC [11]. However, besides HPV status, other risk factors need to be investigated to improve recurrence and survival rates after radiotherapy for patients with HPV(–) HNSCC.
Locoregional recurrence in patients with HPV(–) HNSCC after adjuvant or definitive radiotherapy might be predicted by gene signature as well as clinical or pathological results. So far, the correlation of genetic profiles from some gene sets that perform a role in cancer metabolism with therapeutic response in patients with HNSCC has been investigated [8,12,13]. Also, several studies about association between epithelial-to-mesenchymal transition (EMT) related genes and radioresistance in HNSCC have been reported [14,15]. However, there may be many other molecular biomarker signatures associated with radiosensitivity in patients with HPV(–) HNSCC.
Thus, we aimed to identify and validate radiation sensitivity signature (RSS) of HPV(–) HNSCC from as much gene expression data as possible in order to predict benefits of adjuvant or definitive radiotherapy, which could allow for development of personalized radiotherapy. We hypothesized that investigation of gene expression data and survival assay after irradiation of various HNSCC cell lines would help generate an HNSCC specific RSS, which could be validated in HNSCC big data in The Cancer Genome Atlas (TCGA).
Materials and Methods1. Cell line culturesHNSCC cell lines CAL27, SCC25, and SCC9 were purchased from the American Type Culture Collection (ATCC); HSC2, HSC3, and HSC4 were obtained from the Japanese Cancer Resources Bank (JCRB); and FaDu, SNU1076, SNU1214, SNU46, YD10B, YD38, and YD8 were purchased from the Korean Cell Line Bank (KCLB). The cell line SNU899 was provided by Dr. Kim CH of Ajou University. We used each HNSCC cell line within 3 months of purchase; cell lines used after 3 months of purchase were authenticated and characterized before use. CAL27 cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin (PS). FaDu cells were cultured in Eagle’s Minimum Essential Medium (MEM) supplemented with 10% FBS and 1% PS. SCC25 and SCC9 cells were cultured in DMEM/F-12 supplemented with 10% FBS and 400 ng/mL hydrocortisone. All the other cell lines (HSC2, HSC3, HSC4, SNU1076, SNU1214, SNU46, SNU899, YD10B, YD38, and YD8) were cultured in RPMI supplemented with 10% FBS and 1% PS. The cultures were incubated in a 5% CO2 incubator at 37°C.
2. Clonogenic survival assaysExponentially growing cells were irradiated with 2 Gy radiation using a 250-kVp X-ray (0.61 Gy/min). Then, the irradiated cells were replated in a serum-containing medium at cloning densities. Cells were grown for 10 to 16 days, following which they were fixed and stained with Gentian violet. Colonies of more than 50 cells were scored. The same process was repeated three times for each cell line. The colony-forming efficiency of irradiated cells was determined, averaged, and normalized to that of non-irradiated control cells. The surviving fraction after 2 Gy of radiation (SF-2Gy) was calculated for all 14 cell lines.
3. Investigating RNA expression in cell lines using CCLERNA expression data of each HNSCC cell line was retrieved from the Cancer Cell Line Encyclopedia (CCLE) database (http://www.broadinstitute.org/ccle) [16]. Gene expression data of about 18,361 genes was searched, log transformed, median centered, and scaled to the same minimum/maximum for each HNSCC cell line.
4. Identification of RSSThe survival fraction of 14 HNSCC cell lines after irradiation with 2 Gy radiation was assessed as described above, and four radioresistant cell lines showing high survival fraction values and four radiosensitive cell lines showing low survival fraction values were selected. Receiver operating characteristic (ROC) curve was generated by calculating sensitivity and specificity of expression of each gene in radioresistant and radiosensitive cell lines. Spearman correlation was used to determine the correlation of SF-2Gy with RNA expression data of each HNSCC cell line obtained from the CCLE. Fold change was calculated for every gene between the 25th and 75th percentile of expression. RSS were selected if it satisfied all four inclusion criteria: (1) area under the ROC curve (AUC) ≥0.99, (2) fold change ≥4.3, (3) Spearman correlation R ≥0.6, and (4) Spearman correlation p < 0.05.
5. Patient cohortThis study analyzed the HNSCC cohorts from TCGA. Gene expression levels and clinical data of TCGA were downloaded from the UCSC Cancer Genomics Browser (https://xena.ucsc.edu/public). Patients with HPV(–) TCGA HNSCC who received adjuvant or definitive radiotherapy were selected for validation of radiosensitive genes obtained from HNSCC cell lines. The gene expression profile of TCGA HNSCC cohorts was measured experimentally using the Illumina HiSeq 2000 RNA Sequencing platform at the University of North Carolina TCGA genome characterization center. All the gene expression data of TCGA HNSCC cohorts was log transformed, median centered, and scaled to the same minimum/maximum as in the HNSCC cell lines to make the disparate platforms comparable.
6. Prediction, validation, and statistical analysisUsing data of eight HNSCC cell lines (four radioresistant and four radiosensitive cell lines) as a training set, class prediction procedure was carried out for patients with HPV(–) TCGA HNSCC using compound covariate predictor (CCP) class prediction engine [17,18] with leave-one-out cross-validation (BRB Array Tools) [19]. Genes significantly different between the classes at 0.99 significance level were used for class prediction. Only the genes with a fold-difference between the two classes exceeding 2 were used for class prediction. Then, patients with HPV(–) TCGA HNSCC were classified into radioresistant and radiosensitive groups. Recurrence-free survival (RFS) was defined as the number of months from the date of diagnosis to the event of recurrence [20]. We used Kaplan-Meier method to produce RFS curves in each group of the TCGA cohort. Then, log-rank test was used to compare the RFS between two groups. Univariate and multivariate Cox proportional hazards modeling was performed to evaluated independent prognostic factors associated with the recurrence of HPV(–) HNSCC. p < 0.05 was considered statistically significant. R software package (http://www.r-project.org) was used for all statistical analyses.
7. Pathway analysisThe identified RSS was analyzed by Qiagen’s ingenuity pathway analysis (IPA) software program. At first, the identified RSS was uploaded into Qiagen’s IPA system (http://www.ingenuity.com) for core analysis and then was overlaid with the global molecular network in the ingenuity pathway knowledge base (IPKB) [21]. IPA was performed to identify canonical pathways, upstream regulators, diseases and functions, and gene networks related to radiosensitive genes in HNSCC.
Results1. Development of RSSTo develop radiosensitive genes in HNSCC, we investigated the SF-2Gy of 14 HNSCC cell lines. The SF-2Gy in each cell line ranged from 48% to 72% (Fig. 1A). Then, we identified four radioresistant (FaDu, SNU1076, YD38, HSC3) and four radiosensitive cell lines (SCC9, YD10B, HSC2, SCC25). The value of SF-2Gy was significantly higher in radioresistant cell lines than radiosensitive cell lines (62%–72% vs. 48%–51%; p = 0.028) (Fig. 1A).
Forty-one genes with significant correlation with SF-2Gy were identified in the radioresistant and radiosensitive cell lines through AUC, fold change, and Spearman correlation analysis (Fig. 1B). Of these, 6 genes were positively correlated and 35 genes were negatively correlated with radioresistance, respectively. A full list of these genes is depicted in Supplementary Table S1.
2. Prediction and validation of radiation sensitivity signatureThe gene expression profiles and clinical data of patients with HPV(–) TCGA HNSCC who received adjuvant or definitive radiotherapy were used in this study (n = 203) (Table 1). Class prediction was applied to the data of patients with HPV(–) TCGA HNSCC using BRB Array Tools, following which the HPV(–) TCGA HNSCC cohorts were classified into radioresistant (n = 149) and radiosensitive (n = 54) groups (Fig. 2A). Five-year RFS rate was significantly lower in the radioresistant group than in the radiosensitive group (57.8% vs. 80.1%, respectively; p = 0.035). The corresponding Kaplan-Meier curves are presented in Fig. 2B.
In addition, univariate and multivariate Cox proportional hazards models including patients’ demographics, social history, and clinical staging showed that only RSS is an independent prognostic factor of recurrence in patients with HPV(–) HNSCC who received adjuvant or definitive radiotherapy (p < 0.05) (Table 2).
3. Ingenuity pathway analysisTo identify canonical pathways, upstream regulators, diseases and functions, and gene networks related to radiosensitive genes in HNSCC, RSS was analyzed by Qiagen’s IPA software program. The top canonical pathways, upstream regulators, molecular and cellular functions related to RSS are summarized in Table 3. PDE4B, LILRB4, RARRES3, IFNL4, and TICAM1 were found to be the top upstream regulators of RSS. Moreover, we found that cell death and survival, cell to cell signaling, and cellular movement were significantly enriched in the RSSs. To identify key genes involved in radiosensitivity in HPV(–) HNSCC and establish the connections in-between these genes, the gene network diagram was made based on IPKB and is depicted in Fig. 3. The gene network diagram showed that RSSs were highly correlated with each other.
Discussion and ConclusionIn this study, we developed a molecular biomarker signature of radiation response in HPV(–) HNSCC using various HNSCC cell lines. We classified independent HPV(–) TCGA HNSCC cohort into radioresistant and radiosensitive groups based on RSS. Recurrence after adjuvant or definitive radiotherapy was significantly lower in the radiosensitive group than in the radioresistant group. Thus, RSS in HPV(–) HNSCC may have the ability to identify patients with HNSCC who are refractory to adjuvant or definitive radiotherapy and need treatment intensification or personalized treatment. These RSSs showed various interactive pathways and networks related to cellular movement, cell death and survival, amino acid metabolism, cell morphology, and cell-to-cell signaling.
We used various HPV(–) HNSCC cell lines to include each subsite of HNSCC such as oral cavity, pharynx, supraglottis, glottis, and subglottis in the study. In addition, standard fractionation (2 Gy) was applied to the HNSCC cell lines to evaluate each surviving fraction after irradiation. The mRNA expression levels of each HNSCC cell line were obtained from the CCLE database (http://www.broadinstitute.org/ccle), because CCLE may provide representative genetic proxies for primary tumors in many cancer types [22].
Moreover, an enormous amount of biological and clinical data—big data—facilitated us to validate RSSs from the HNSCC cell lines. TCGA is the largest dataset and contains measurements of somatic mutations (sequencing), copy number variations (array based and sequencing), mRNA expression (array based and sequencing), miRNA expression (array based and sequencing), protein expression (array based), and histology slides for approximately 7,000 human tumors (http://cancergenome.nih.gov/). Here, from the clinical data of 604 TCGA HNSCC patients, the data of 203 patients with HPV(–) TCGA HNSCC who received radiotherapy was selected for validation.
We could interpret the gene expression profiles using advanced bioinformatic analysis as revealed by microarray. In this study, we used the IPA software program—one of the advanced bioinformatic analysis tools—which can analyze the gene expression patterns using a built-in scientific literature based database (www.ingenuity.com) [21]. IPA results helped us in finding cellular functions and several pathways and networks related to RSS. It has been reported that five top upstream regulators, namely, PDE4B, LILRB4, RARRES3, IFNL4, TICAM1, are associated with colorectal cancer [23], leukemia [24], breast cancer [25], prostate cancer [26], and thyroid cancer [27], respectively. These regulators seem to be associated with radiosensitivity in some ways. The levels of intracellular cAMP, which is a well-characterized secondary messenger that elicits a wide range of cellular processes including proliferation, differentiation, migration, growth, and apoptosis, are regulated by the activities of two enzymes, adenylyl cyclase and phosphodiesterase including PDE4B [23]. Appukuttan et al. [28] found that inhibition of soluble adenylyl cyclase had increased the radiosensitivity of prostate cancer cells. Interferon-related DNA damage resistance signature (IRDS) induced by all interferon including IFNL4 showed effects leading to resistance to ionizing radiation [26]. Association between T lymphocyte and other regulators such as LILRB4, TICAM1 had been reported [24,29]. These regulators are not RSSs of HPV(–) HNSCC, but needed to be further investigated for their association with HNSCC and radiotherapy. In addition, various molecular and cellular functions were enriched in the RSS of HPV(–) HNSCC and gene network framework showed that the RSSs were highly correlated with each other in a complex manner.
Recently, some studies have investigated the correlation of genetic mechanisms and treatment response including radiotherapy in patients with HNSCC. MRP2 and RB were found to be associated with the outcome of concurrent chemoradiation in patients with HNSCC [12]. Additionally, the expression levels of cancer stem cell markers CD44, SLC3A2, and MET were correlated with tumor recurrence in HPV(–) HNSCC after postoperative chemoradiation [13]. Several hallmarks including EMT which refers to a process whereby the adhesive polarity of epithelial cancer cells dissipates and changes to mesenchymal cells, angiogenesis, and DNA repair were significantly correlated with 13 radioresistance-associated genes [14]. In a recent study, a seven-gene signature predicting recurrence in patients with locally advanced HNSCC treated with postoperative radiotherapy or radiochemotherapy was developed and validated [8]. These results were developed from gene sets well known for their role in biological or cancer metabolism such as DNA repair, cancer stem cells, chemoresistance, HPV association, hypoxia, proliferation, etc. Conversely, this study developed several novel RSSs from HNSCC cell lines using CCLE regardless of the role of the genes.
There had been also some reports showing association between the RSSs and radiosensitivity. TXNRD1, which was significantly increased in radioresistant group, encodes thioredoxin reductase 1 (TrxR1). Several studies reported that specific inhibition of TrxR1 increased radiosensitivity via enhancement ROS levels [30-32]. Luo et al. [33] found that PDLIM4 gene was involved in radioresistance of glioma cells. PDLIM4 necessary for dendritic cell migration via CCR7-JNK, dendrite formation, and subsequent development of functional T-cell responses was inactivated in multiple cancer types [34]. ALDH2 detoxifies toxic aldehydes formed by accumulated reactive oxygen resulted from ionizing radiation. Ning et al. [35] reported that reactive aldehydes played an important role in the intrinsic radiosensitivity of normal and tumor tissue. The chemokine CXCL10 plays a role in angiostasis and has anti-tumor effects. It was reported that high expression of CXCL10 was related to better response to neoadjuvant chemoradiotherapy as well as improved survival in colorectal cancer patients [36,37]. Lukas et al. [38] found that a young woman with ROS1 oncogene rearranged non-small cell lung cancer (NSCLC) with brain metastases was sensitive to radiotherapy. PDLIM4, ALDH2, CXCL10, and ROS1 were significantly decreased in radioresistant group. However, there are still many RSSs that need to be investigated for their association with radiosensitivity in carcinoma, especially HNSCC.
Our study showed that only RSS was an independent prognostic factor of RFS in patients with HPV(–) TCGA HNSCC who received adjuvant or definitive radiotherapy. Clinical tumor stage was not prognostic factor in TCGA dataset. Most patients (n = 176) were diagnosed with advanced stage and only 20 patients were diagnosed with early stage, because early stage patients requiring only surgical therapy were excluded in this study. Also, among 20 early stage patients, 11 patients received operation with radiotherapy, and 3 patients received operation with chemoradiotherapy. In other words, most patients in early stage needed additional therapy, maybe because of their relative advanced state. We think these characteristics might influence the RFS in patients with HPV(–) HNSCC who received adjuvant or definitive radiotherapy.
Our study has the following limitations. First, the subsites of HNSCC cell lines are diverse and include oral cavity, pharynx, and larynx. Thus, diverse radiation sensitivity of each subsite might have been overlooked. However, we focused on overall HPV(–) HNSCC which has poor prognosis than HPV(+) HNSCC. Second, we did not isolate RNA from the HNSCC cell lines, but used mRNA expression data from CCLE. Since CCLE might provide data with genomic similarities to primary tumors [22], we used HNSCC cell lines within three months of purchase or used them after authentication. Finally, little is known about the RSSs developed in this study. Further experiments including IPA described above are needed to comprehend the underlying molecular mechanism of each RSS.
Nevertheless, as far as we know, this is the first study to identify RSSs associated with HPV(–) HNSCC from gene database of various HNSCC cell lines. We investigated patients with HPV(–) HNSCC treated by adjuvant or definitive radiotherapy irrespective of operation, focusing on their prognosis that is worse than that in patients with HPV(+) HNSCC. In addition, RSSs were validated using big data—patients with HPV(–) TCGA HNSCC treated by radiotherapy. It means that our validation results for RSSs might be influenced by minimal selective error and could be reproducible. Furthermore, Cox proportional hazards model showed that only RSS is the independent prognostic factor influencing recurrence of HPV(–) HNSCC in patients after radiotherapy. As each patient had HNSCC at a different clinical stage and was treated with different amount of radiation dose, a direct comparison between radioresistant and radiosensitive groups might be impractical. However, the sole prognostic factor, RSS, needs to be noted and further investigated.
In conclusion, we derived 41 RSSs that could predict recurrence in HPV(–) HNSCC. The outcome of adjuvant or definitive radiotherapy in HPV(–) HNSCC can be predicted by analyzing the expression of RSSs. Further validation of RSS according to each subsite of HNSCC in other HNSCC cohorts may facilitate the application of RSSs in treatment. In other words, personalized treatment plan for treatment of HPV(–) HNSCC could be established by further organization and stratification of RSSs.
AcknowledgmentsThis research was supported by the Bio & Medical Technology Development Program of the National Research Foundation (NRF), funded by the Korean government (MSIP&MOHW) (No. NRF-2018R1D1A1B07050154). The study was also supported by a grant from Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (No. HI18C1039).
Supplementary MaterialsSupplementary materials can be found via https://doi.org/10.3857/roj.2020.00136.
Table S1.Genes correlated with survival fraction after 2-Gy radiation in HPV-negative head and neck squamous cell carcinoma cell lines. Fig. 1.Clonogenic survival assays performed to assess the surviving fraction after 2 Gy of radiation (SF-2Gy) identified the range of radioresistance in 14 HPV(–) HNSCC cell lines (48%–72%). (A) Four radioresistant cell lines and four radiosensitive cell lines were selected based on SF-2Gy in each HPV(–) HNSCC cell line (62%–72% vs. 48%–51%; p = 0.028). On comparison of the radioresistant and radiosensitive cell lines, 41 genes with significant correlation with radioresistance were identified; 6 genes were positively correlated with radioresistance and 35 genes were negatively correlated with radioresistance. (B) Relative gene expression levels of these 41 genes in the four radioresistant and four radiosensitive cell lines are depicted. HPV, human papillomavirus; HNSCC, head and neck squamous cell carcinoma. 
Fig. 2.Using four radioresistant and four radiosensitive cell lines with 41 genes significantly correlated with radioresistance as a training set, class prediction was carried out for patients with HPV(–) TCGA HNSCC who received adjuvant or definitive radiotherapy. (A) The patients with HPV(–) TCGA HNSCC were classified into radioresistant (n = 149) and radiosensitive (n = 54) groups. (B) Using Kaplan-Meier curves, 5-year RFS rate of each group was determined. The radioresistant group showed significantly lower 5-year RFS rate than the radiosensitive group (57.8% vs. 80.1%; p = 0.035). HPV, human papillomavirus; TCGA, The Cancer Genome Atlas; HNSCC, head and neck squamous cell carcinoma; RFS, recurrence-free survival. 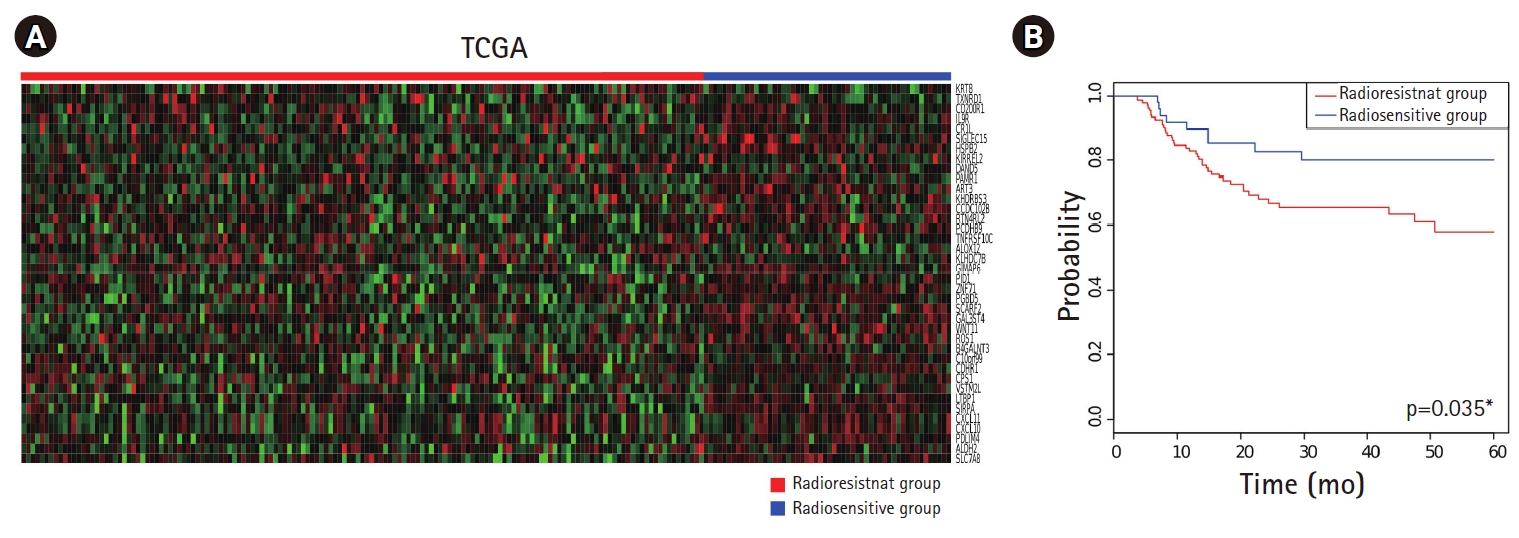
Fig. 3.Gene network diagram based on ingenuity pathway analysis showing connections in-between radiation sensitivity signatures. 
Table 1.Patients’ characteristics (n = 203) Table 2.Univariate and multivariate analysis of factors associated with recurrence-free survival in HPV(–) head and neck squamous cell carcinoma patients who received adjuvant or definitive radiotherapy
Table 3.Top canonical pathways, upstream regulators, molecular and cellular functions related to radiation sensitivity signature References1. Kamangar F, Dores GM, Anderson WF. Patterns of cancer incidence, mortality, and prevalence across five continents: defining priorities to reduce cancer disparities in different geographic regions of the world. J Clin Oncol 2006;24:2137–50.
3. Leemans CR, Braakhuis BJ, Brakenhoff RH. The molecular biology of head and neck cancer. Nat Rev Cancer 2011;11:9–22.
4. Smith RB, Sniezek JC, Weed DT, Wax MK; Microvascular Surgery Subcommittee of American Academy of Otolaryngology-Head and Neck Surgery. Utilization of free tissue transfer in head and neck surgery. Otolaryngol Head Neck Surg 2007;137:182–91.
5. Vergeer MR, Doornaert PA, Rietveld DH, Leemans CR, Slotman BJ, Langendijk JA. Intensity-modulated radiotherapy reduces radiation-induced morbidity and improves health-related quality of life: results of a nonrandomized prospective study using a standardized follow-up program. Int J Radiat Oncol Biol Phys 2009;74:1–8.
6. Gupta S, Kong W, Peng Y, Miao Q, Mackillop WJ. Temporal trends in the incidence and survival of cancers of the upper aerodigestive tract in Ontario and the United States. Int J Cancer 2009;125:2159–65.
7. Gujral DM, Nutting CM. Patterns of failure, treatment outcomes and late toxicities of head and neck cancer in the current era of IMRT. Oral Oncol 2018;86:225–33.
8. Schmidt S, Linge A, Zwanenburg A, et al. Development and validation of a gene signature for patients with head and neck carcinomas treated by postoperative radio(chemo)therapy. Clin Cancer Res 2018;24:1364–74.
9. Ragin CC, Taioli E. Survival of squamous cell carcinoma of the head and neck in relation to human papillomavirus infection: review and meta-analysis. Int J Cancer 2007;121:1813–20.
10. Lohaus F, Linge A, Tinhofer I, et al. HPV16 DNA status is a strong prognosticator of loco-regional control after postoperative radiochemotherapy of locally advanced oropharyngeal carcinoma: results from a multicentre explorative study of the German Cancer Consortium Radiation Oncology Group (DKTK-ROG). Radiother Oncol 2014;113:317–23.
11. Gupta AK, Lee JH, Wilke WW, et al. Radiation response in two HPV-infected head-and-neck cancer cell lines in comparison to a non-HPV-infected cell line and relationship to signaling through AKT. Int J Radiat Oncol Biol Phys 2009;74:928–33.
12. van den Broek GB, Wildeman M, Rasch CR, et al. Molecular markers predict outcome in squamous cell carcinoma of the head and neck after concomitant cisplatin-based chemoradiation. Int J Cancer 2009;124:2643–50.
13. Linge A, Lock S, Gudziol V, et al. Low cancer stem cell marker expression and low hypoxia identify good prognosis subgroups in HPV(-) HNSCC after postoperative radiochemotherapy: a multicenter study of the DKTK-ROG. Clin Cancer Res 2016;22:2639–49.
14. Foy JP, Bazire L, Ortiz-Cuaran S, et al. A 13-gene expression-based radioresistance score highlights the heterogeneity in the response to radiation therapy across HPV-negative HNSCC molecular subtypes. BMC Med 2017;15:165.
15. de Jong MC, Ten Hoeve JJ, Grenman R, et al. Pretreatment microRNA expression impacting on epithelial-to-mesenchymal transition predicts intrinsic radiosensitivity in head and neck cancer cell lines and patients. Clin Cancer Res 2015;21:5630–8.
16. Goldman M, Craft B, Swatloski T, et al. The UCSC Cancer Genomics Browser: update 2015. Nucleic Acids Res 2015;43(Database issue):D812–7.
17. Radmacher MD, McShane LM, Simon R. A paradigm for class prediction using gene expression profiles. J Comput Biol 2002;9:505–11.
18. Ramaswamy S, Tamayo P, Rifkin R, et al. Multiclass cancer diagnosis using tumor gene expression signatures. Proc Natl Acad Sci U S A 2001;98:15149–54.
19. Simon R, Lam A, Li MC, Ngan M, Menenzes S, Zhao Y. Analysis of gene expression data using BRB-ArrayTools. Cancer Inform 2007;3:11–7.
20. Ding D, Stokes W, Eguchi M, et al. Association between lymph node ratio and recurrence and survival outcomes in patients with oral cavity cancer. JAMA Otolaryngol Head Neck Surg 2019;145:53–61.
21. Yu J, Gu X, Yi S. Ingenuity pathway analysis of gene expression profiles in distal nerve stump following nerve injury: insights into Wallerian degeneration. Front Cell Neurosci 2016;10:274.
22. Barretina J, Caponigro G, Stransky N, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012;483:603–7.
23. Kim DU, Kwak B, Kim SW. Phosphodiesterase 4B is an effective therapeutic target in colorectal cancer. Biochem Biophys Res Commun 2019;508:825–31.
24. Deng M, Gui X, Kim J, et al. LILRB4 signalling in leukaemia cells mediates T cell suppression and tumour infiltration. Nature 2018;562:605–9.
25. Anderson AM, Kalimutho M, Harten S, Nanayakkara DM, Khanna KK, Ragan MA. The metastasis suppressor RARRES3 as an endogenous inhibitor of the immunoproteasome expression in breast cancer cells. Sci Rep 2017;7:39873.
26. Tang W, Wallace TA, Yi M, et al. IFNL4-ΔG allele is associated with an interferon signature in tumors and survival of African-American men with prostate cancer. Clin Cancer Res 2018;24:5471–81.
27. Sigurdson AJ, Brenner AV, Roach JA, et al. Selected single-nucleotide polymorphisms in FOXE1, SERPINA5, FTO, EVPL, TICAM1 and SCARB1 are associated with papillary and follicular thyroid cancer risk: replication study in a German population. Carcinogenesis 2016;37:677–84.
28. Appukuttan A, Flacke JP, Flacke H, Posadowsky A, Reusch HP, Ladilov Y. Inhibition of soluble adenylyl cyclase increases the radiosensitivity of prostate cancer cells. Biochim Biophys Acta 2014;1842(12 Pt B):2656–63.
29. Shime H, Matsumoto M, Seya T. Double-stranded RNA promotes CTL-independent tumor cytolysis mediated by CD11b+Ly6G+ intratumor myeloid cells through the TICAM-1 signaling pathway. Cell Death Differ 2017;24:385–96.
30. Liang YW, Zheng J, Li X, Zheng W, Chen T. Selenadiazole derivatives as potent thioredoxin reductase inhibitors that enhance the radiosensitivity of cancer cells. Eur J Med Chem 2014;84:335–42.
31. Selenius M, Hedman M, Brodin D, et al. Effects of redox modulation by inhibition of thioredoxin reductase on radiosensitivity and gene expression. J Cell Mol Med 2012;16:1593–605.
32. Smart DK, Ortiz KL, Mattson D, et al. Thioredoxin reductase as a potential molecular target for anticancer agents that induce oxidative stress. Cancer Res 2004;64:6716–24.
33. Luo M, Cai W, Li K, Liu J, Xue W, Liu Y. Effect of PDLIM4 gene on the prognosis of glioma and its cell radiosensitivity. Zhonghua Fang She Yi Xue Yu Fang Hu Za Zhi (Chin J Radiol Med Prot) 2017;37:725–9, 762.
34. Yoo JY, Jung NC, Lee JH, et al. Pdlim4 is essential for CCR7-JNK-mediated dendritic cell migration and F-actin-related dendrite formation. FASEB J 2019;33:11035–44.
35. Ning S, Budas GR, Churchill EN, Chen CH, Knox SJ, Mochly-Rosen D. Mitigation of radiation-induced dermatitis by activation of aldehyde dehydrogenase 2 using topical alda-1 in mice. Radiat Res 2012;178:69–74.
36. Jiang Z, Xu Y, Cai S. CXCL10 expression and prognostic significance in stage II and III colorectal cancer. Mol Biol Rep 2010;37:3029–36.
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||

 |
 |