Introduction
Carcinoembryonic antigen (CEA) was first identified in 1965 [1]. It is also known as CD66 or CEA-related cell adhesion molecule 5 [2] and has a molecular mass of 180–200 kDa depending on the extent of its glycosylation [3]. It is a member of a large family composed of 29 genes divided into three subgroups, including CEA-like glycoproteins and pregnancy-specific glycoproteins, all of which are members of the much larger immunoglobulin supergene family [4,5].
CEA is the most widely used tumor marker for colorectal cancer (CRC). In 2000, the American Joint Committee on Cancer stated that preoperative elevation of the serum CEA level is an independent prognostic factor for CRC [6]. In addition to its prognostic role, the predictive role of CEA has recently received much attention [7]. Since the standard-of-care for locally advanced rectal cancer changed from postoperative chemoradiotherapy to preoperative chemoradiotherapy, the tumor response of each patient can be identified early on by surgical pathology. Adjusted approaches, such as conservative local excision or no surgery, are currently under clinical investigation for selected patients with an excellent tumor response [8,9]. Many reports have shown that serum CEA levels are significantly associated with this chemoradiation response [7]. Research on the molecular mechanisms related to CEA would aid our understanding of how CEA plays both a prognostic and predictive role in CRC.
To identify CEA-interacting proteins, we applied a proteomics approach using liquid chromatographytandem mass spectrometry (LC-MS/MS) analysis of CEA immunoprecipitates in human colon cancer cell lines. After irradiation, changes in the expression of the identified CEAinteracting proteins were monitored by Western blot analysis.
Materials and Methods
1. Human colon cancer cell lines
Human colon cancer cell lines SNU-81 and LoVo were obtained from the Korean Cell Line Bank (Seoul, Korea).
2. Immunoprecipitation
Immunoprecipitation was performed as described previously [10]. All procedures were performed at 4°C unless otherwise specified. Approximately 107 cells in 1 mL cold 1 × RIPA buffer containing protease inhibitors (Roche Diagnostics, Basel, Switzerland) were incubated on ice for 30 minutes with occasional mixing. Cell lysates were centrifuged at 12,000 × g for 10 minutes, and the supernatant was carefully collected without disturbing the pellet. The supernatant was mixed with the CEA primary antibody and incubated for 2 hours on a rocking platform. Prepared Protein G Sepharose beads (100 μL; GE Healthcare Life Sciences, Marlborough, MA, USA) were added, followed by further incubation on ice for 1 hour on a rocking platform. The mixture was centrifuged at 10,000 × g for 30 seconds, and the supernatant was removed completely. Protein G Sepharose beads were washed five times with 1 mL cold 1 × RIPA buffer to minimize the background. Next, 100 μL 2 × SDS sample buffer were added to the bead pellet and heated to 100°C for 10 minutes. After boiling, the immunoprecipitates were centrifuged at 10,000 × g for 5 minutes, and the supernatant was collected for Western blot analysis.
3. Mass spectrometry analysis
Mass spectrometry (MS) analysis was performed as described previously [11]. Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) gels containing the proteins of interest were excised, destained with 50% acetonitrile in 0.1 M ammonium bicarbonate, and dried in a SpeedVac evaporator. Dried gel pieces were re-swollen with 30 μL 25 mM sodium bicarbonate, pH 8.8, containing 50 ng trypsin (Promega, Madison, WI, USA) at 37°C overnight. Samples were desalted using Zip-Tips C18 (Millipore, Bedford, MA, USA) and dissolved in 10 μL 2% acetonitrile in 0.1% formic acid. MS analysis was performed using a LTQ XL linear ion trap mass spectrometer system (Thermo Fisher Scientific, San Jose, CA, USA) at the Proteomics Core, National Cancer Center, Korea. The mass spectrometer was set for nanospray ionization (NSI) in positive mode. A syringe pump was used to introduce the calibration solution for automatic tuning and calibration of the LTQ in NSI positive ion mode. Infusion of digested samples (trypsin) into the ionization source of the mass spectrometer was accomplished by liquid chromatographic separation. The spray voltage was set at +1.1 kV, the temperature of the capillary was set at 200°C, the capillary voltage was set at +20 V, and the tube lens voltage was set at +100 V. The auxiliary gas was set to zero. Full-scan experiments were performed to linear trap in the range m/z 150–2,000. Systematic MS/MS experiments were performed by changing the relative collision energy and monitoring the intensities of the fragment ions. All MS/MS samples were analyzed using Sequest (Thermo Fisher Scientific; version v.27, rev. 11). Sequest was set up to search the uniprot_sprot database and the IPI human database assuming that the digestion enzyme was trypsin. Sequest was searched using a fragment ion mass tolerance of 1.00 Da and a parent ion tolerance of 1.2 Da. Oxidation of methionine was specified in Sequest as a variable modification.
4. Irradiation of human colon cancer cells and Western blot analysis
The X-RAD 320 Biological Irradiator (Precision X-Ray, North Branford, CT, USA) was used to deliver precise radiation dosages (2, 4, and 6 Gy) to human colon cancer cells. After irradiation, Western blot analysis was performed as described previously [10]. Briefly, equal amounts of protein were subjected to SDS-PAGE. Following electrophoresis, proteins were transferred onto polyvinylidene fluoride membranes (Millipore) and blocked by overnight incubation at 4°C. Membranes were incubated with a primary antibody against CEA (Abcam, Cambridge, MA, USA), lysosome C (Abcam), Rab-6B (Abnova, Atlanta, GA, USA) or β-actin (Santa Cruz Biotechnology, Dallas, TX, USA). Membranes were washed and incubated with a diluted horseradish peroxidase-conjugated secondary antibody (Southern Biotech, Birmingham, AL, USA). Membranes were washed again (3 × 15 minutes), incubated with WEST-ZOL chemiluminescence reagent (iNtRON Biotechnology, Seoul, Korea) for 1 minutes, and exposed to Blue XB-1 film (Kodak, Rochester, NY, USA).
5. Cell proliferation assay post-irradiation
A colorimetric assay using the tetrazolium salt MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide) was performed to assess cell proliferation after irradiation (X-RAD 320 biological irradiator). MTT assays were performed as described previously [11].
Results
1. CEA expression in human colon cancer cell lines and CEA detection in immunoprecipitates
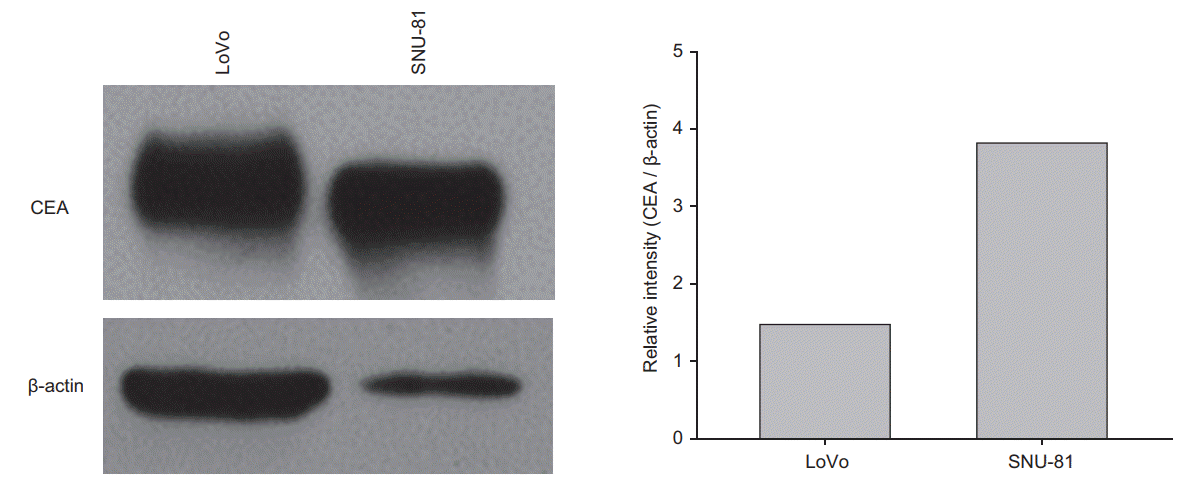
CEA expression was confirmed in human colon cancer cell lines (LoVo and SNU-81) by Western blot analysis. After normalization to β-actin, CEA expression was higher in SNU-81 cells compared with LoVo cells (Fig. 1). Using the anti-CEA antibody [IP(CEA)] or IgG immunoprecipitates as a negative control, immunoprecipitation was performed in whole cell lysates (WL) from each cell line. WL, as a positive control for CEA, and IgG and IP(CEA) immunoprecipitates were loaded onto the SDS-PAGE gel for Western blot analysis. An immunoreactive signal for CEA was detected in the WL and IP(CEA) but not in the IgG immunoprecipitates (Supplementary Fig. S1).
2. Identification of CEA-interacting proteins by proteome analysis of the anti-CEA antibody immunoprecipitates
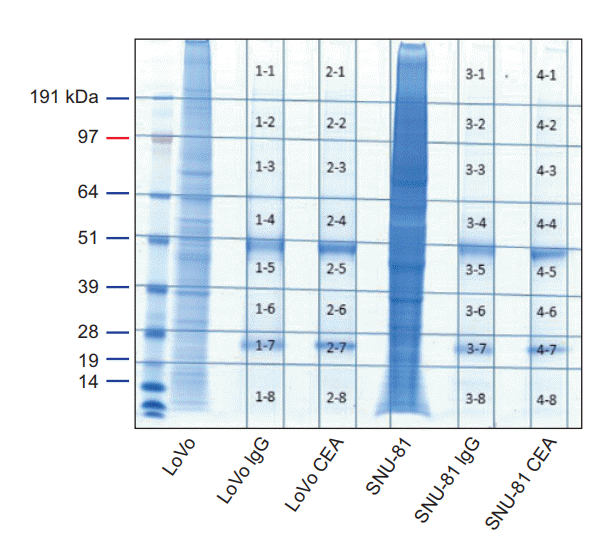
The protein samples used in Fig. 1B were loaded again onto SDS-PAGE gels, and gel staining was performed as shown in Fig. 2. The stained gel was then sliced into 32 pieces (1-1 to 4-8 in Fig. 2) and used for LC-MS/MS analysis. The identified proteins are listed in Supplementary Tables S1 to S4. CEAinteracting proteins in LoVo (or SNU-81) cells were obtained by excluding the proteins identified in slices 1-1 to 1-8 (or 3-1 to 3-8) from the proteins identified in slices 2-1 to 2-8 (or 4-1 to 4-8).
The Ras-related protein Rab-6B and lysozyme C were identified as CEA-interacting proteins in LoVo and SNU-81 cells, respectively (Table 1). No CEA-interacting protein was identified in both cell lines.
3. Effect of irradiation on the expression of CEA, Rab-6B and lysozyme C
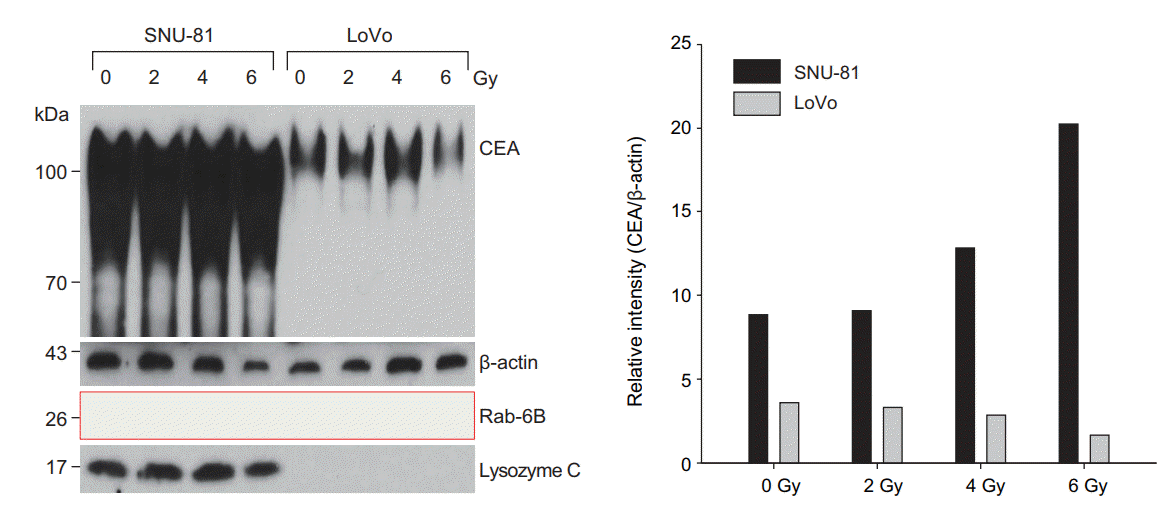
The expression of CEA was higher in SNU-81 cells compared with LoVo cells (Figs. 1, 3). CEA expression after irradiation was differently regulated in two cell lines; CEA expression in SNU-81 cells was increased after irradiation, but it was decreased in LoVo cells in radiation dosage-dependent manner (Fig. 3). The expression of lysosome C was not affected by irradiation and was detected only in SNU-81 cells (Fig. 3). This finding may explain why lysozyme C was identified as a CEA-interacting protein only in SNU-81 cells (Fig. 3, Table 1). Commercially available antibody against Rab-6B was not capable to detect Rab-6B expression (Fig. 3). Presence of Rab-6B in LoVo cells was confirmed by immunoprecipitate using anti-CEA antibody (Supplementary Fig. S2).
4. Differential radiation response of SNU-81 and LoVo cells
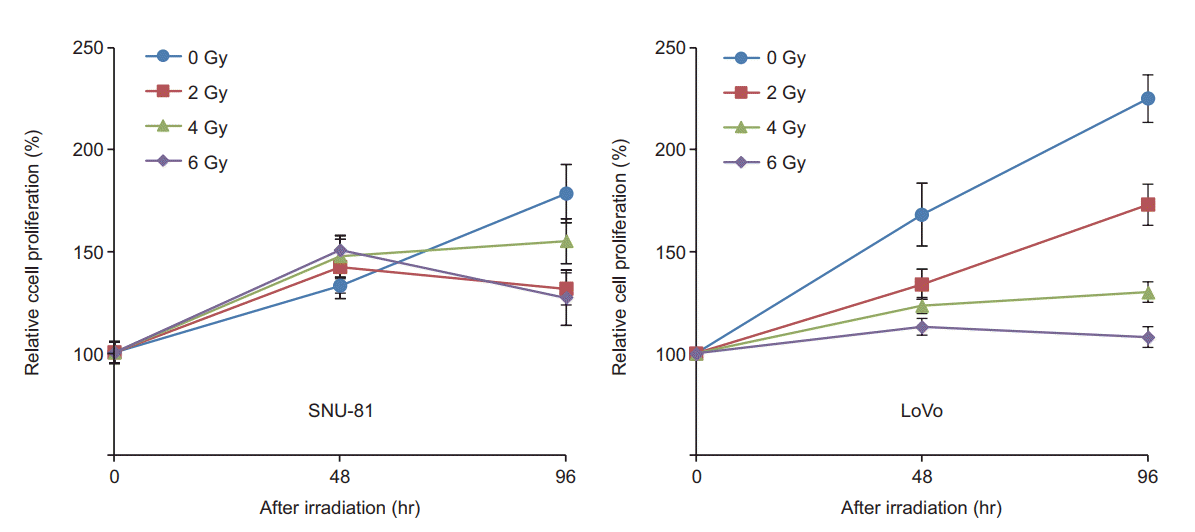
After irradiation (2, 4, and 8 Gy), the proliferation rates of SNU-81 and LoVo cells were determined by MTT assay. As shown in Fig. 4, the proliferation rate of LoVo cells was higher than that of SNU-81 cells but was more suppressed by irradiation in a dosage-dependent manner.
Discussion and Conclusion
All CEA family members function as intercellular adhesion molecules [12]. There is abundant evidence that CEA is involved in multiple biological aspects of cancer, such as cell adhesion, cell signaling, immune mechanisms, apoptosis, anoikis, angiogenesis and metastasis [13-17]. For example, CEA increased the ability of CRC cells to colonize in the liver or lung [18,19]. CEA significantly protected HT29 CRC cells from undergoing apoptosis under various conditions, including treatment with 5-fluorouracil [14]. However, the mechanism by which elevated CEA is related to the unfavorable radiation susceptibility of CRC cells remains to be elucidated.
The present study aimed to identify CEA-interacting proteins. Proteomic analysis of immunoprecipitates using an anti-CEA antibody can provide many candidate molecules that interact with CEA and may reveal the currently unknown molecular function of CEA by classifying CEA-interacting proteins. However, we identified only two CEA-interacting proteins, Rab-6B and lysozyme C, in LoVo and SNU-81 cells, respectively. At this moment, we are unable to explain the reason why only two proteins were identified. However, the subcellular localization of CEA may provide a clue; CEA is localized in the cellular membrane, which may limit the number of proteins that immunoprecipitate with CEA.
Both cell lines showed very different expression patterns. The expression of CEA (normalized to β-actin) was much higher in SNU-81 cells compared with LoVo cells (Figs. 1, 3). CEA expression was down-regulated in LoVo cells but it was increased in SNU-81 cells in a radiation dosage-dependent manner (Fig. 3). Furthermore, the proliferation rate of LoVo cells was more suppressed by irradiation in a dosagedependent manner (Fig. 4). These findings suggest that downregulation of CEA expression may increase the sensitivity of the cells to radiation. However, this should be further verified by employing various types of CRC cell lines and a CEA suppression technique, such as siRNA or shRNA.
The detection of lysosome C expression only in SNU-81 cells explained why lysozyme C was identified as a CEAinteracting protein in these cells only (Fig. 3, Table 1). Lysozyme is one of the major protein components of human milk, but it is also synthesized by various types of cancer cells [20,21]. The expression of lysozyme was positively correlated with a favorable outcome in breast cancer [20] and was upregulated to a greater extent in mucinous cell carcinomas than in well-differentiated and poorly differentiated gastric adenocarcinomas [21]. Lysozyme directly activates immune cells and increases tumor cell immunogenicity [22], which have been confirmed in a number of experimental cases [22-24]. However, even without immune cell activation, self-assembled nanostructured hen egg white lysozyme and recombinant human lysozyme showed strong antiproliferative effects in breast [23] and gastric [24] cancer cells, respectively. However, a better understanding of the antitumor mechanisms of lysozyme (intracellular or extracellular) is needed. The commercially available lysozyme antibody used in this study detected all subtypes of lysozyme, including type C, but failed to detect lysozyme C in LoVo cells (Fig. 3). Lysozyme C appeared to be highly down-regulated or absent in LoVo cells. However, further studies are needed to clarify whether lysozyme expression affects the radiation response in CRC cells.
Unfortunately, we failed to detect Rab-6B using the commercially available antibody (Fig. 3). We assumed that the endogenous expression of Rab-6B was as much lower as unable to detect with Western blot analysis. However, interaction between CEA and Rab-6B was confirmed in IP using anti-CEA antibody and whole cell lysate of LoVo cells (Supplementary Fig. S2). Various roles of Rab family proteins in cancer have been reported. Rab-1 is a mediator of dynamic membrane trafficking between the endoplasmic reticulum and Golgi membrane, which plays an important role in cellular signaling transduction and in the synthesis and transport of proteins and lipids [25]. Rab-1 dysregulation has been reported in cancer [25]. The Rab GDP dissociation inhibitor beta may play a role as a tumor suppressor, and its expression is downregulated in non-small cell lung cancer [26]. Moreover, the expression levels of Rab and Rab effector genes are decreased in bladder cancer [27]. As a tumor suppressor, miR-200b regulates the expression of Rab family proteins in breast cancer [28]. Rab 5C-dependent endocytosis and glycolysis are down-regulated in cisplatin-resistant ovarian cancer [29]. With regard to radiation, Rab-14 shows an association in nasopharyngeal carcinoma. The expression of Rab-14 is suppressed by miR-451, which results in an increased radiation response in nasopharyngeal carcinoma by inhibiting the repair of irradiation-induced double-strand breaks and increasing apoptosis [30]. However, the association between Rab-6B, which was determined to interact with CEA in the present study, and radiation sensitivity remains to be elucidated.
This study has limitations. Rectal cancer cell lines were not used in the experiments because they are known to show very high resistance to chemotherapy and radiation compared with colon cancer cells [31]. LoVo and SNU-81 cells were chosen among colon cancer cell lines because they show relatively high expression of CEA, and each of them are derived from Caucasian and Korean, respectively. The endogenous expression of lysozyme C and radiation-induced down-regulation of CEA differed between two cell lines. The CEA-mediated radiation response may vary depending on the characteristics of individual cancer cells.
In conclusion, LC-MS/MS analysis of CEA immunoprecipitates in LoVo and SNU-81 cells was used to identify Rab-6B and lysozyme C as CEA-interacting proteins, respectively. This study suggests that lysozyme C and Rab subfamily proteins may play a role in the relationship between CEA and radiosensitivity of CRC cells, although further studies are needed to clarify functional roles of the identified proteins.